NCERT Solutions for Class 12 Biology Chapter 11 Biotechnology : Principles and Processes
Q1 : Can you list 10 recombinant proteins which are used in medical practice? Find out where they are used as therapeutics (use the internet).
Answer :
Recombinant proteins are obtained from the recombinant DNA technology. This technology involves the transfer of specific genes from an organism into another organism using vectors and restriction enzymes as molecular tools.
Ten recombinant proteins used in medical practice are –
| Recombinant protein | Therapeutic use |
| Insulin | Treatment for type I diabetes mellitus |
| Interferon-α | Used for chronic hepatitis C |
| Interferon -μ² | Used for herpes and viral enteritis |
| Coagulation factor VII | Treatment of haemophilia A |
| Coagulation factor IX | Treatment of haemophilia B |
| DNAase I | Treatment of cystic fibrosis |
| Anti-thrombin III | Prevention of blood clot |
| Interferon B. | For treatment of multiple sclerosis |
| Human recombinant growth hormone | For promoting growth in an individual |
| Tissue plasminogen activator | Treatment of acute myocardial infection |
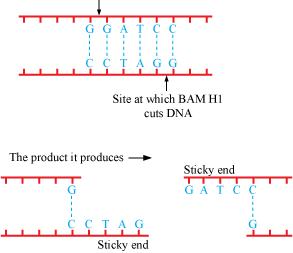
Q2 : Make a chart (with diagrammatic representation) showing a restriction enzyme, the substrate DNA on which it acts, the site at which it cuts DNA and the product it produces.
Answer :
The name of the restriction enzyme is Bam H 1.
Q3 : From what you have learnt, can you tell whether enzymes are bigger or DNA is bigger in molecular size? How did you know?
Answer :
Enzymes are smaller in size than DNA molecules. This is because DNA contains genetic information for the development and functioning of all living organisms. It contains instructions for the synthesis of proteins and DNA molecules. On the other hand, enzymes are proteins which are synthesised from a small stretch of DNA known as ‘genes’, which are involved in the production of the polypeptide chain.
Q4 : What would be the molar concentration of human DNA in a human cell? Consult your teacher.
Answer :
The molar concentration of human DNA in a human diploid cell is as follows:
⇒ Total number of chromosomes x 6.023 x 1023
⇒ 46 x 6.023 x 1023
⇒ 2.77 x 1018 Moles
Hence, the molar concentration of DNA in each diploid cell in humans is 2.77 x 1023 moles.
Q5 : Do eukaryotic cells have restriction endonucleases? Justify your answer.
Answer :
No, eukaryotic cells do not have restriction endonucleases. This is because the DNA of eukaryotes is highly methylated by a modification enzyme, called methylase. Methylation protects the DNA from the activity of restriction enzymes .These enzymes are present in prokaryotic cells where they help prevent the invasion of DNA by virus.
Q6 : Besides better aeration and mixing properties, what other advantages do stirred tank bioreactors have over shake flasks?
Answer :
The shake flask method is used for a small-scale production of biotechnological products in a laboratory. On the other hand, stirred tank bioreactors are used for a large-scale production of biotechnology products.
Stirred tank bioreactors have several advantages over shake flasks:
(1) Small volumes of culture can be taken out from the reactor for sampling or testing.
(2) It has a foam breaker for regulating the foam.
(3) It has a control system that regulates the temperature and pH.
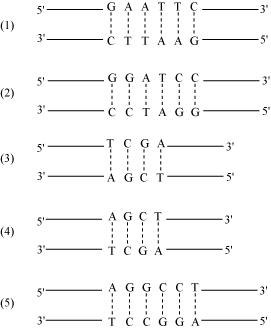
Q7 : Collect 5 examples of palindromic DNA sequences by consulting your teacher. Better try to create a palindromic sequence by following base-pair rules.
Answer :
The palindromic sequence is a certain sequence of the DNA that reads the same whether read from 5′ → 3′ direction or from 3′ → 5′ direction. They are the site for the action of restriction enzymes. Most restriction enzymes are palindromic sequences.
Five examples of palindromic sequences are:
Q8 : Can you recall meiosis and indicate at what stage a recombinant DNA is made?
Answer :
Meiosis is a process that involves the reduction in the amount of genetic material. It is two types, namely meiosis I and meiosis II. During the pachytene stage of prophase I, crossing over of chromosomes takes place where the exchange of segments between non-sister chromatids of homlogous chromosomes takes place. This results in the formation of recombinant DNA.
Q9 : Can you think and answer how a reporter enzyme can be used to monitor transformation of host cells by foreign DNA in addition to a selectable marker?
Answer :
A reporter gene can be used to monitor the transformation of host cells by foreign DNA. They act as a selectable marker to determine whether the host cell has taken up the foreign DNA or the foreign gene gets expressed in the cell. The researchers place the reporter gene and the foreign gene in the same DNA construct. Then, this combined DNA construct is inserted in the cell. Here, the reporter gene is used as a selectable marker to find out the successful uptake of genes of interest (foreign genes). An example of reporter genes includes lac Z gene, which encodes a green fluorescent protein in a jelly fish.
Q10 : Describe briefly the followings:
(a) Origin of replication
(b) Bioreactors
(c) Downstream processing
Answer :
(a) Origin of replication – Origin of replication is defined as the DNA sequence in a genome from where replication initiates. The initiation of replication can be either uni-directional or bi-directional. A protein complex recognizes the ‘on’ site, unwinds the two strands, and initiates the copying of the DNA.
(b) Bioreactors – Bioreactors are large vessels used for the large-scale production of biotechnology products from raw materials. They provide optimal conditions to obtain the desired product by providing the optimum temperature, pH, vitamin, oxygen, etc. Bioreactors have an oxygen delivery system, a foam control system, a PH, a temperature control system, and a sampling port to obtain a small volume of culture for sampling.
(c) Downstream processing – Downstream processing is a method of separation and purification of foreign gene products after the completion of the biosynthetic stage. The product is subjected to various processes in order to separate and purify the product. After downstream processing, the product is formulated and is passed through various clinical trials for quality control and other tests.
Q11 : Explain briefly: (a) PCR (b) Restriction enzymes and DNA (c) Chitinase
Answer :
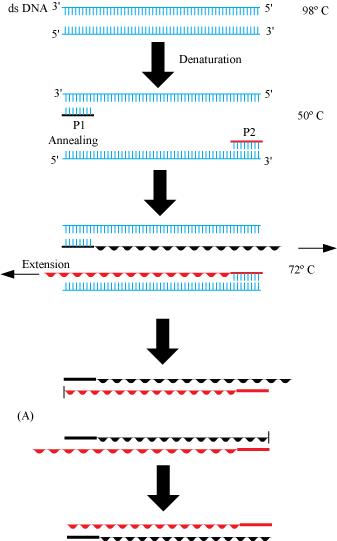
(a) PCR: – Polymerase chain reaction (PCR) is a technique in molecular biology to amplify a gene or a piece of DNA to obtain its several copies. It is extensively used in the process of gene manipulation. The process involves in-vitro synthesis of sequences using a primer, a template strand, and a thermostable DNA polymerase enzyme obtained from a bacterium, called Thermus aquaticus. The enzyme utilizes building blocks dNTPs (deoxynucleotides) to extend the primer. In the first step, the double stranded DNA molecules are heated to a high temperature so that the two strands separate into a single stranded DNA molecule. This process is called denaturation. Then, this ssDNA molecule is used as a template strand for the synthesis of a new strand by the DNA polymerase enzyme and this process is called annealing, which results in the duplication of the original DNA molecule. This process is repeated over several cycles to obtain multiple copies of the rDNA fragment.
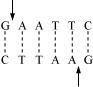
(b) Restriction enzymes and DNA: Restriction enzymes are molecular scissors used in molecular biology for cutting DNA sequences from a specific site. It plays an important role in gene manipulation. The enzymes recognize a specific six-box pair sequence known as the recognition sequence and cut the sequence at a specific site. For example, the recognition site for enzyme ECORI is as follows:
Restriction enzyme are categorized into two types –
(i) Exonuclease – It is a type of restriction enzyme that removes the nucleotide from either 5′ or 3′ ends of the DNA molecule.
(ii) Endonuclease – It is a type of restriction enzyme that makes a cut within the DNA at a specific site. This enzyme acts as an important tool in genetic engineering. It is commonly used to make a cut in the sequence to obtain DNA fragments with sticky ends, which are later joined by enzyme DNA ligase.
(c) Chitinase – Chitinase is a class of enzymes used for the degradation of chitin, which forms a major component of the fungal cell wall. Therefore, to isolate the DNA enclosed within the cell membrane of the fungus, enzyme chitinase is used to break the cell for releasing its genetic material.
Q12 : Discuss with your teacher and find out how to distinguish between
(a) Plasmid DNA and chromosomal DNA
(b) RNA and DNA
(c) Exonuclease and Endonuclease
Answer :
(a) Plasmid DNA and chromosomal DNA
| Plasmid DNA | Chromosomal DNA |
| Plasmid DNA is an extra-chromosomal DNA molecule in bacteria that is capable of replicating, independent of chromosomal DNA. | Chromosomal DNA is the entire DNA of an organism present inside chromosomes. |
(b) RNA and DNA
| RNA | DNA |
| RNA is a single stranded molecule. | DNA is a double stranded molecule. |
| It contains ribose sugar. | It contains deoxyribose sugar. |
| The pyrimidines in RNA are adenine and uracil. | The pyrimidines in DNA are adenine and thymine. |
| RNA cannot replicate itself. | DNA molecules have the ability to replicate. |
| It is a component of the ribosomes. | It is a component of the chromosomes. |
(c) Exonuclease and Endonuclease
| Exonuclease | Endonuclease |
| It is a type of restriction enzyme that removes the nucleotide from 5′ or 3′ ends of the DNA molecule. | It is a type of restriction enzyme that makes a cut within the DNA at a specific site to generate sticky ends. |
